Reproducable research
I have a terrible memory for what I’ve done in the past. I can barely remember what I had for breakfast let alone specifics about a data analysis I did from the past week. I tend to go down so many wrong paths that by the time my weekly supervisor meeting rolls around I have no idea what I’ve done or why I’ve done it. I would often stare dumbfounded when asked seemingly simple questions like, “how did you make this graph?” or even “why did you make this graph?”
So, for the past few months I’ve started producing weekly reports using R and multimarkdown to give to my supervisors before our meetings. It’s a summary of my work from the past week. This basically constitutes a lab notebook for me. A lot of my work involves writing a lot of math formulae, producing plots and images, some code to munge data, and text to explain what I’m doing.
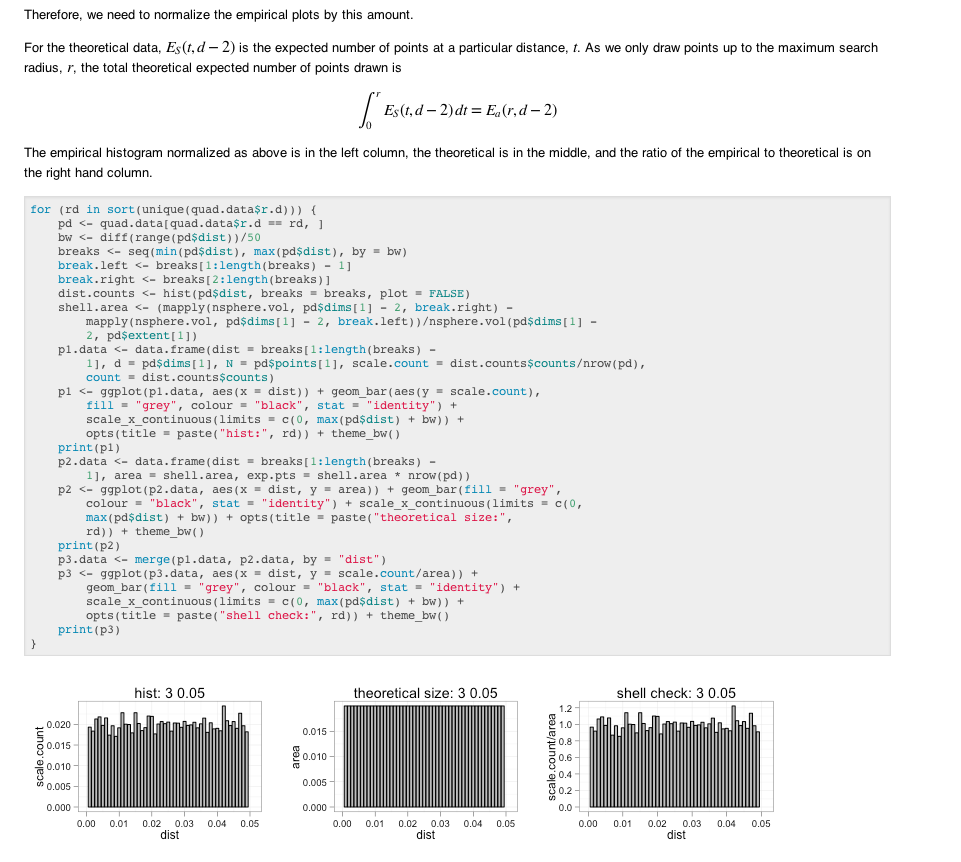
I use R for any sort of numerical computing and multimarkdown has been my go-to markup language for some time. My favorite feature of multimarkdown is its support for processing LaTeX math. Now that the knitr package is out I can tie my analysis code (written in R) to my explanations (written in markdown). What’s also great about this system is that my analysis is contained entirely in a text file that I can store in source control along with any data and the rest of my project.
The rest of this post goes into detail about how I’ve set up Vim, knitr, and Marked.app on my Mac to automatically update my report every time I save. The code is located at https://github.com/gabysbrain/r-notebook.
Editor: Vim
The best thing about this method is that it’s pretty much editor agnostic. Marked doesn’t care what editor you use as long as it’s editing markdown. So, I get to use the only text editor I’ve ever liked: vim!
I linked Marked to vim with the instructions from A Whole lot of Bollocks’ blog post so that I can hit <leader>-m and have the file I’m editing open in Marked. Once the file is open it automatically updates every time I save.
To make my editing more efficient I use the SnipMate plugin which brings TextMate-style snippets to vim. Basically, you can type a shorter string, hit tab, and have it expand into anything you want. It’s really great for adding boilerplate code. I particularly like this for adding the knitr code blocks. With the fork of SnipMate I have set up (https://github.com/gabysbrain/snipmate.vim) typing rcode expands to
<!--begin.rcode ${1},cache=TRUE,dependson='${2}'
${3}
end.rcode-->and typing rfig expands to
<!--begin.rcode ${1},cache=TRUE,dependson='${2}',fig.width=${3:12},fig.height=${4:12},dev='png'
${5}
end.rcode-->I’m also using the multimarkdown syntax highlighter: https://github.com/jngeist/vim-multimarkdown.
Processor: knitr
In its default incarnation knitr provides a lot of functionality out of the box for processing R code embedded in a markdown document. The killer feature (other than the markdown output) in my opinion is its caching and and dependency layer so I don’t have to wait for R to recompute every section when I just change one. It’s also smart enough to recompute any blocks that depend on the one I changed.
The main issue is that knitr is designed to be run from the desired working directory. This is how it sets the locations for the images and cache files. Luckily, when you run a custom markdown processor in recent versions of Marked it appends the directory containing whatever file you’re editing to the end of the path. So, all you have to do before running code through knitr is change the current working directory to the last item in PATH and the cache and images will be in the proper place.
This script also allows me to add some custom hooks into the markdown processor. I mostly use this to test out new features before submitting them to the main knitr development.
Viewer: Marked
This all gets piped through Marked. Marked.app is an amazing and simple markdown preview tool. When the preview is open it watches my source file for changes and automatically updates when I save in vim.
By default it uses its own multimarkdown processor but you can override that with your own. It also supports a custom css file. The one I’m using is slightly modified from the github theme but with nicer tables.
Future
Right now I print the final document to PDF and then email the file to my supervisors. Eventually though I’m going to upload the whole thing to a private research blog that we all share.
I’d also like to add a lot more interactive features to the HTML. The killer features would be interactive manipulation of graphics and having those changes reflected in the source document. It would also be great if I could inspect R objects directly in the HTML document as well. This would require some interaction with a running version of R though.
But for now, I’m happy that I can easily make a record of what I’ve done each week and my supervisor is happy that I can show what I’ve done!